The reshape function
reshape function. I will focus on the reshape function in base R, and not the package of the same name.I use Fischer’s iris data set again, as it is readily available after starting R. The iris data set has 150 observation and the first 6 rows look like this:
data(iris)
head(iris)## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosaI would like to create a box whisker plot, showing the measurements of the observations for each of the species, as in the chart below.
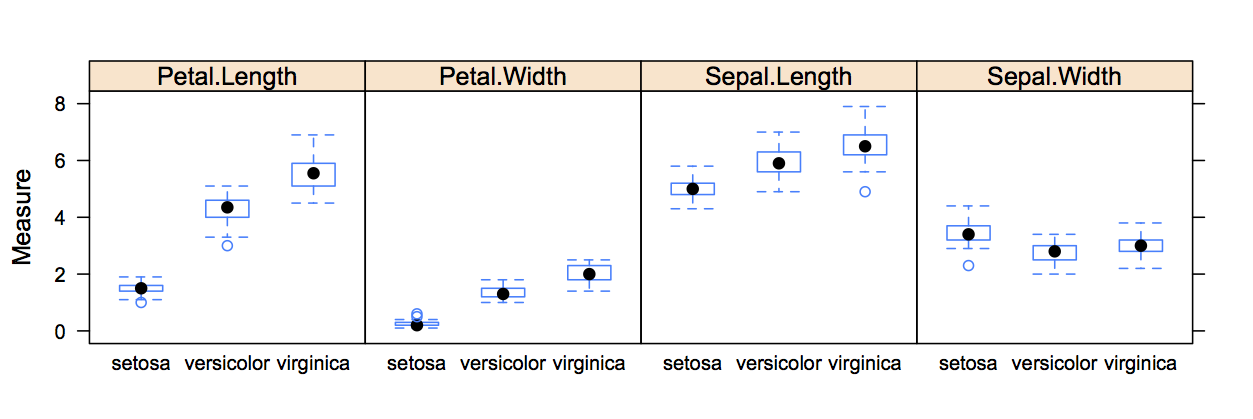
I know, that if I had all measurements in one column and the dimension in another column, I could produce a graph like this in one line with lattice.
library(lattice)
bwplot(Measurement ~ Species | Dimension, data=reshaped.iris)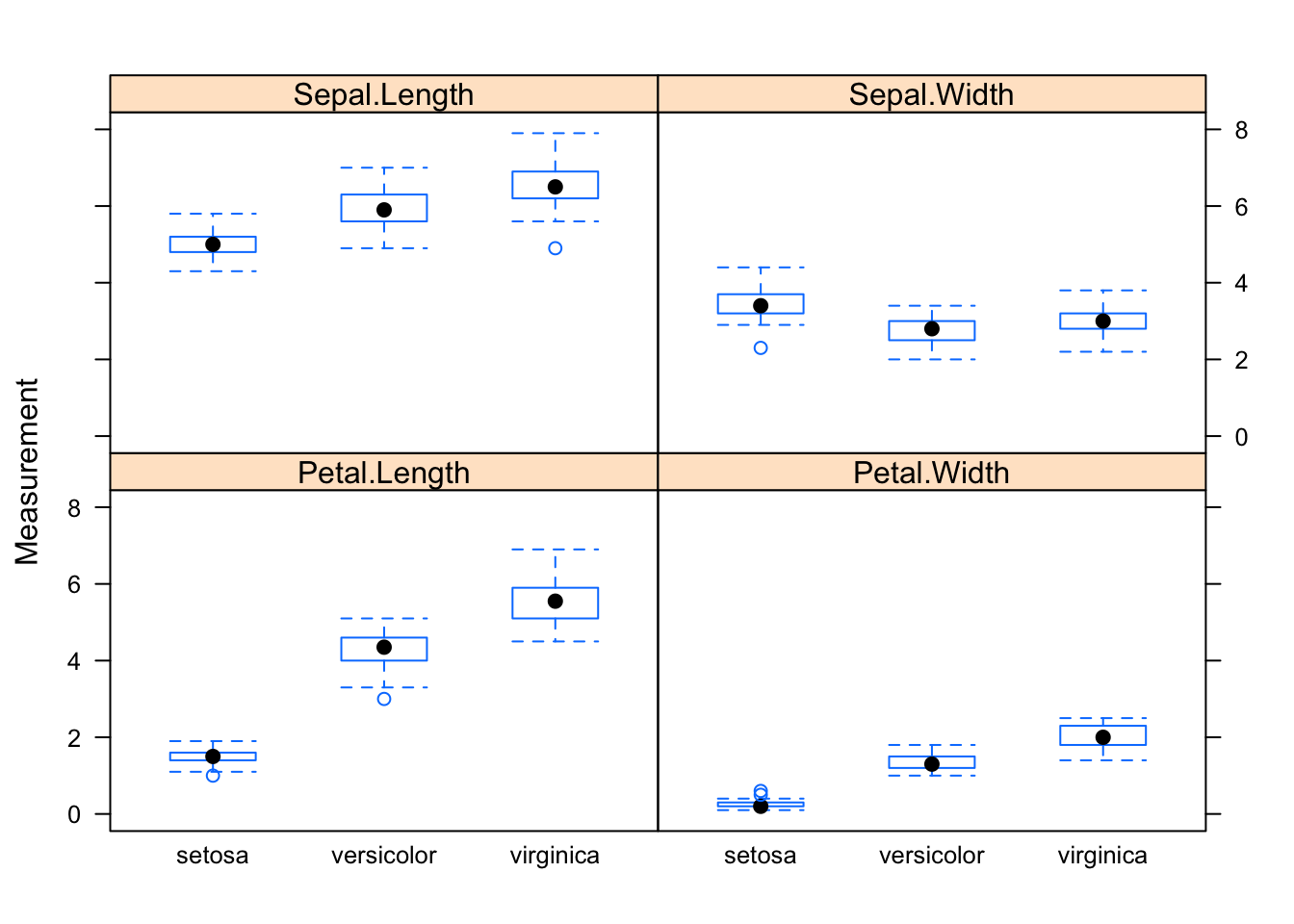
Hence the reshape function is what I need. From the help file I learn that I want to transform my data from a wide format into a long format (direction=“long”). In the long format I would like a varibale with the measurements (v.names=“Measurement”), which I get by running through the first four columns (varying=1:4). I know which measurement I am reading by looking at the column names (times=names(iris)[1:4]), and I capture the dimension names in a new variable (timevar=“Dimension”). This gives me the following statement:
reshaped.iris <- reshape(iris, varying=1:4, v.names="Measurement",
timevar="Dimension", times=names(iris)[1:4],
idvar="Measure ID", direction="long")
head(reshaped.iris)## Species Dimension Measurement Measure ID
## 1.Sepal.Length setosa Sepal.Length 5.1 1
## 2.Sepal.Length setosa Sepal.Length 4.9 2
## 3.Sepal.Length setosa Sepal.Length 4.7 3
## 4.Sepal.Length setosa Sepal.Length 4.6 4
## 5.Sepal.Length setosa Sepal.Length 5.0 5
## 6.Sepal.Length setosa Sepal.Length 5.4 6That’s it, I can create the lattice box-whisker plot.
In my next example I would like the measurements of length and width in separate columns and capture the flower part in a new variable, so I can create scatterplots of length against width. Tweaking the reshape statement slightly gives me:
reshaped.iris.sp <- reshape(iris, varying=list(c(1,3),c(2,4)),
v.names=c("Length", "Width"),
timevar="Part", times=c("Sepal", "Petal"),
idvar="Measure ID", direction="long")
head(reshaped.iris.sp)## Species Part Length Width Measure ID
## 1.Sepal setosa Sepal 5.1 3.5 1
## 2.Sepal setosa Sepal 4.9 3.0 2
## 3.Sepal setosa Sepal 4.7 3.2 3
## 4.Sepal setosa Sepal 4.6 3.1 4
## 5.Sepal setosa Sepal 5.0 3.6 5
## 6.Sepal setosa Sepal 5.4 3.9 6xyplot(Length ~ Width | Species, groups=Part,
data=reshaped.iris.sp, auto.key=list(space="right"))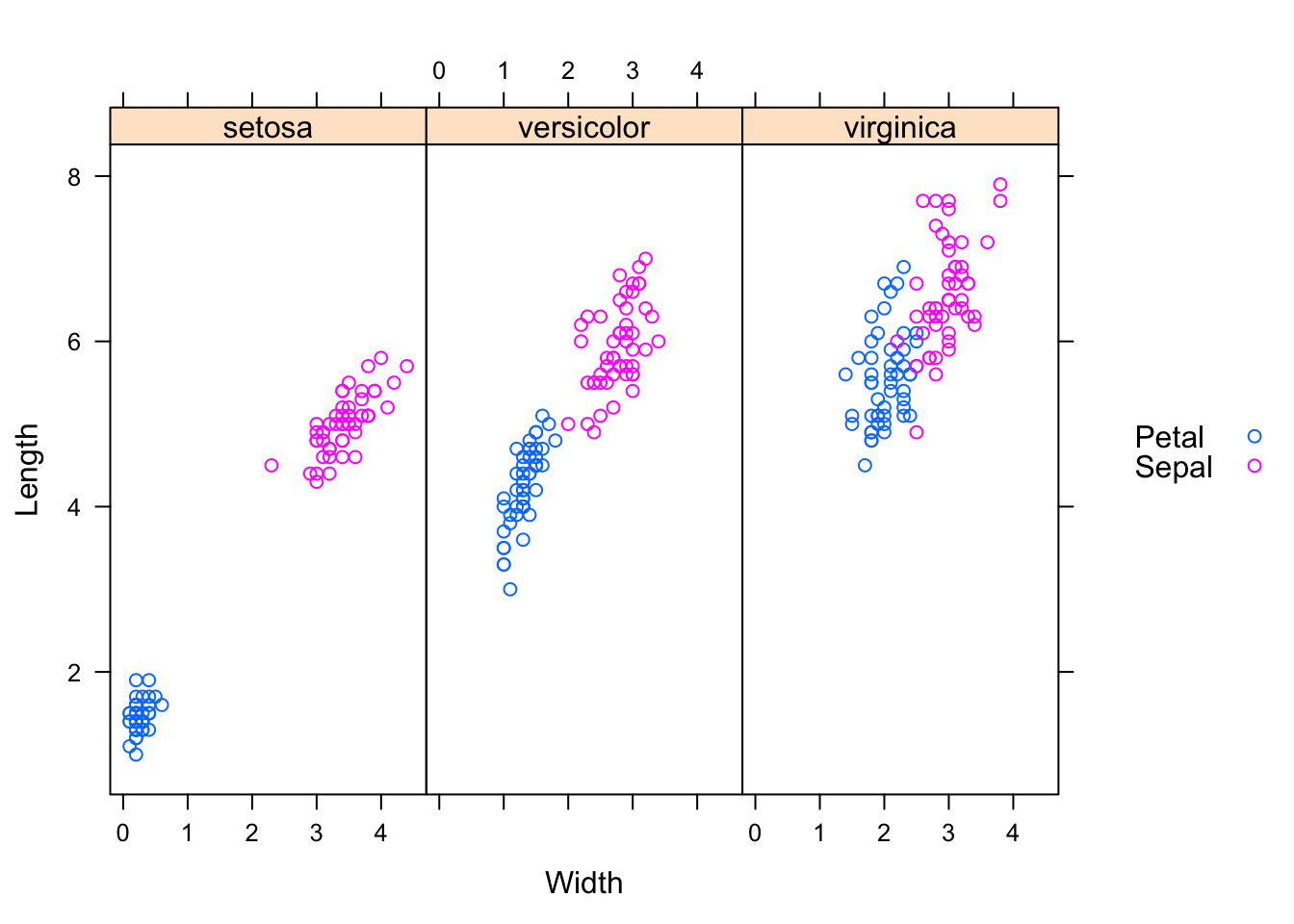
Let’s swap Part against Species.
xyplot(Length ~ Width | Part, groups=Species,
data=reshaped.iris.sp, auto.key=list(space="right"))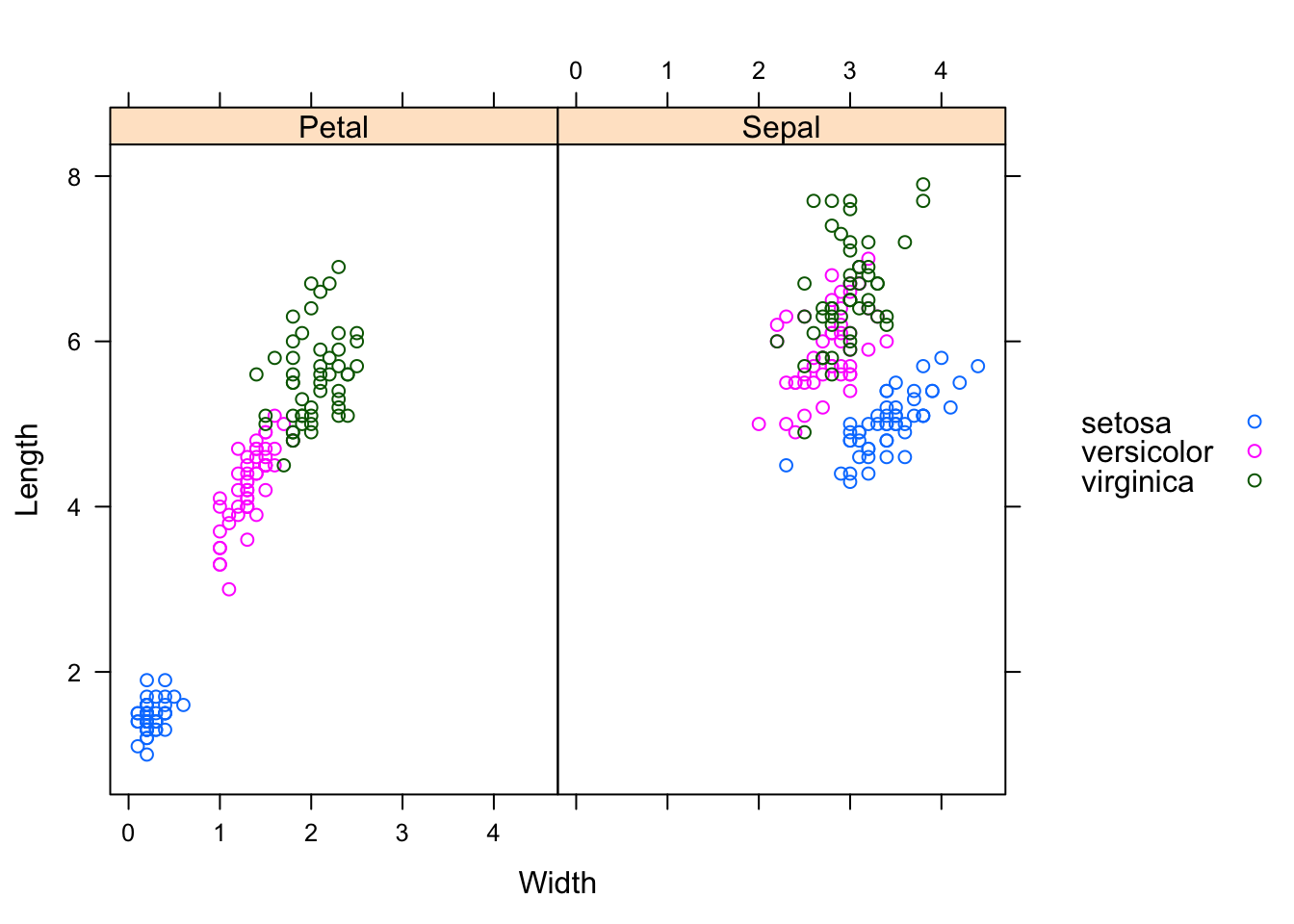
I think, the charts illustrate quite nicely why the iris data set has become a typical test case for many classification techniques in machine learning.
Citation
For attribution, please cite this work as:Markus Gesmann (Feb 09, 2012) The reshape function. Retrieved from https://magesblog.com/post/2012-02-09-reshape-function/
@misc{ 2012-the-reshape-function,
author = { Markus Gesmann },
title = { The reshape function },
url = { https://magesblog.com/post/2012-02-09-reshape-function/ },
year = { 2012 }
updated = { Feb 09, 2012 }
}